
Your Health Magazine
4201 Northview Drive
Suite #102
Bowie, MD 20716
301-805-6805

More Health Technology Articles
CRISPR Screens: How Scientists Find the Genes That Matter
If biology had a “search” button, researchers would press it every day. Why does one tumor shrug off a drug while another responds? Which genes quietly help a virus replicate? What makes a cell age faster—or stay resilient longer?
For a long time, answering questions like these meant inching forward one gene at a time. That’s slow, expensive, and a bit like trying to find one faulty wire by testing every wire in a building. CRISPR changed the pace. Most people know CRISPR as a tool for editing DNA—those famous “molecular scissors.” But in many labs, CRISPR’s most useful superpower isn’t editing one gene. It’s testing thousands of genes in parallel to see which ones actually matter.

That approach is called a CRISPR screen, and it’s one of the most practical ways scientists turn biological chaos into a clear shortlist of causes.
What is CRISPR screening, in normal-people language?
A CRISPR screen is a big, organized experiment where you nudge lots of genes—sometimes across the whole genome—and watch what happens. Instead of asking, “Does gene X affect this?” you ask, “Which genes affect this, period?”
Here’s the basic flow. You start with a large population of cells. You introduce CRISPR guides so that different cells get different genetic changes. Then you apply a challenge: a drug, an infection, heat stress, low nutrients—whatever matches the real-world question. After that, you measure which cells thrive and which struggle. By reading the “barcode” of the CRISPR guides inside the surviving (or failing) cells, you can work backward and identify the genes that helped or hurt under that condition.
It’s like running thousands of tiny A/B tests at once—except the “A/B” is a gene being altered or left alone.
Why people outside science should care
Even if you never plan to step into a lab, CRISPR screens ripple into everyday life because they help speed up discovery in places that touch us all.
- Finding better drug targets: Screens can reveal which genes a disease state truly depends on—so researchers spend less time chasing the wrong suspects.
- Explaining resistance: Treatments fail because cells adapt. A screen can spotlight the escape routes, which is often the first step toward smarter drug combinations.
- Mapping how cells work: Screens help uncover networks—genes that act like main switches, and genes that quietly serve as backups.
In other words: screens help scientists stop guessing and start prioritizing.
The engine behind a screen: guide RNA libraries
CRISPR screens usually rely on a guide RNA library. Think of a library as a carefully designed collection of CRISPR “instructions,” each one aimed at a specific gene. Some libraries are genome-wide. Others focus on a theme—like immune pathways, cancer-related genes, or druggable enzymes. And sometimes researchers build custom libraries to test a shortlist they already suspect.
When you run a screen, you’re essentially asking the library to “try” thousands of genetic tweaks across the cell population and see which tweaks change the outcome.
If you’re exploring how this works end-to-end—especially in the context of designing experiments around phenotypes like survival, sensitivity, or resistance—this overview of crispr screening is a helpful reference because it frames screening as a practical discovery workflow rather than a buzzword.
Pooled vs. arrayed screens: two flavors, two strengths
Not all screens look the same. Two common formats are pooled and arrayed.
Pooled screens are the high-throughput workhorse. All the guides are mixed together and delivered as one big batch. Each cell gets (ideally) one guide. After the challenge, you sequence the guide barcodes to see which ones became more common (those changes helped cells survive) and which ones disappeared (those changes hurt). Pooled screens shine when your readout is something like survival, growth rate, or general “fitness.”
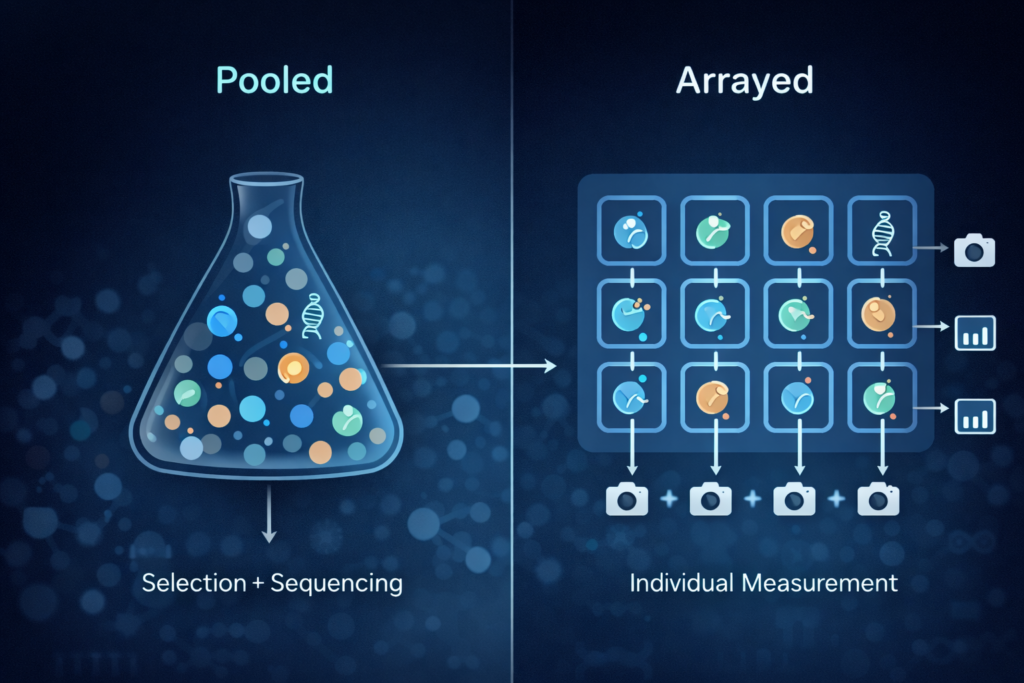
Arrayed screens are more like a tidy spreadsheet. Each well (or sample) gets one guide, so you can measure each perturbation separately. That makes arrayed formats great for complex readouts—like imaging or detailed pathway measurements—but they’re usually more resource-heavy.
A common pattern is pooled for discovery, then follow-up experiments to confirm the best hits.
CRISPR isn’t only about breaking genes
Another reason screens are so flexible is that CRISPR can be used in different “modes,” depending on the question.
- Knockout screens aim to disrupt a gene so it stops working.
- CRISPRi screens dial a gene down (repression) without cutting DNA.
- CRISPRa screens dial a gene up (activation), useful when “more of a gene” drives a trait.
This matters because biology doesn’t always behave well with simple on/off switches. Some genes are essential: fully knocking them out kills the cell, making it hard to learn anything. In those cases, tuning expression up or down can reveal effects that a hard knockout can’t.
What makes a screen trustworthy (and what can go wrong)
CRISPR screens can look deceptively simple on paper, but the quality usually comes down to a few fundamentals:
- Enough coverage: each guide needs to be represented in enough cells, or randomness can masquerade as a “hit.”
- Solid controls: non-targeting guides and known positive controls help show whether the screen behaved as expected.
- A meaningful challenge: selection pressure has to create real separation, not subtle drift.
- Careful analysis: sequencing counts aren’t conclusions; statistics is what turns readouts into confident gene-level signals.
When those pieces are in place, the results can be remarkably clear—and genuinely useful.
Why some teams use screening services
A full CRISPR screen is a chain: library choice and design, delivery, maintaining proper coverage, running the selection, sequencing, and interpreting results. Each stage comes with its own quality-control challenges. That’s why many labs outsource part—or sometimes all—of the workflow, especially if they don’t run screens on a regular basis.
In practice, research teams often work with specialized providers that support CRISPR screening across multiple stages of this process. Ubigene, for example, takes an integrated approach to CRISPR library screening, combining flexible library design with phenotypic screening and structured data readout to help streamline the path from experimental question to interpretable results.
If you’re evaluating what “end-to-end” support can look like, a crispr library screen service typically packages practical pieces like library options, delivery formats, QC checkpoints, sequencing, and reporting—helpful when you want reproducible results without building a full screening pipeline from scratch.
The big takeaway
CRISPR screens are one of the fastest ways scientists can turn a messy biological question into a focused set of answers. They don’t replace careful follow-up experiments—but they do something priceless: they point attention toward the few genes that truly drive an outcome, saving months (or years) of guessing.
And that’s why, behind so many headlines about new therapies, new targets, and new biological insights, there’s often a quieter engine humming in the background: the CRISPR screen.
Other Articles You May Find of Interest...
- CRISPR Screens: How Scientists Find the Genes That Matter
- Women’s Health Apps: How Custom Healthcare Software Is Transforming FemTech Innovation
- How Personalized Fitness and Nutrition Apps Are Transforming Health Outcomes in 2026
- Top HealthTech Trends to Watch in 2026
- The Growing Demand for Stem Cell Treatment in Chicago: What Patients Need to Know
- 5 Steps To A Healthy And Secure Digital Wellness Routine
- The Digital Immune System: How SeqOps and Bangalore’s Cybersecurity Leaders Are Protecting Healthcare in 2026














